MeanShift Clustering: a GPU case-study
What are GPUs again?
A simple way to conceptualise the difference between CPUs, the main processor of a modern computer, and GPUs, the processors on graphics cards, is to think of a CPU as containing one (or few) very fast computing unit while a GPU contains very many, much slower computing units.
For example, my Macbook has a quad-core CPU with ‘hyperthreading’, so four x 2 = 8 computing units each clocked at 3.1 GHz (approximately!). It also has a Radeon Pro graphics card, which contains 1024 computing units each running at 907 MHz. The reason for this very different design gets at the basic difference in their purposes: a CPU is trying to perform a sequence of operations – running your software – as quickly as possible. A GPU is trying to fill all the pixels in your screen, preferably at the same time.
The CPU’s job is largely serial, whereas the GPU’s can all be done in parallel.
Massive parallelism in scientific computing
Very often, mathematical or scientific computations involve large numbers of independent operations. For example, you may want to perform a calculation on multiple data, or samples in a dataset, or agents in a population. Or perhaps you want to process the same data, but using different parameters in your operations. All of these cases may be called ‘massively parallel’, because they represent a large number of computations that are all independent, and hence can all be performed at the same time on different computing units.
This is sometimes also called ‘embarrassingly parallel’, since almost no extra work need be done to parallelise such computations.
MeanShift Clustering:1 massive parallelism on the GPU
MeanShift is a non-parametric2 algorithm for finding maxima in the density functions of distributions. Given some discrete samples (your dataset), MeanShift estimates the modes (peaks) in the density profile of the sample, as follows:
For each point i in the sample, compute the weighted sum J(i) of all of its neighbours j within some neighbourhood of i.
- This neighbourhood is defined by a kernel function, which is often simply a Gaussian centred on i and with sigma set to a parameter called the Bandwidth.
Replace i <- J(i). This shifts i to the centre of mass of all points in its neighbourhood.
Continue this process until i converges onto J(i) (ie, the shifts become negligible). These limiting points are the modes of the density distribution!
Label each point i with the limiting J(i) to which its mean shifts converged.
If we assume that a set of data contains clusters, and that the samples will be most dense at the centres of these clusters, then the above algorithm allows us to find them by letting the set of labels {J(i)} define the cluster membership.
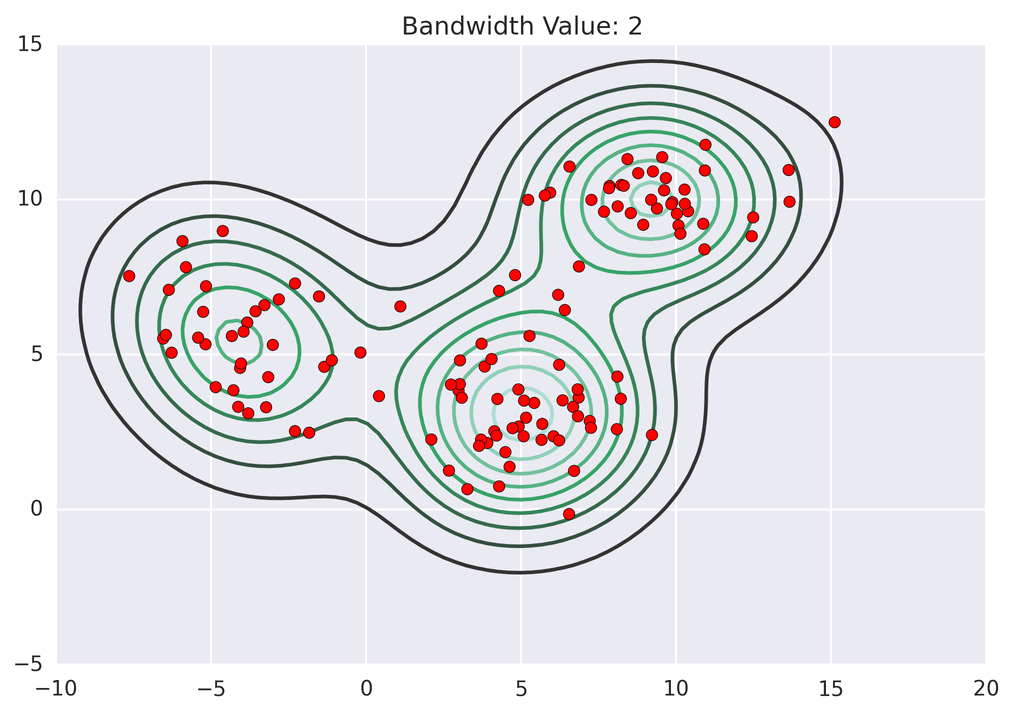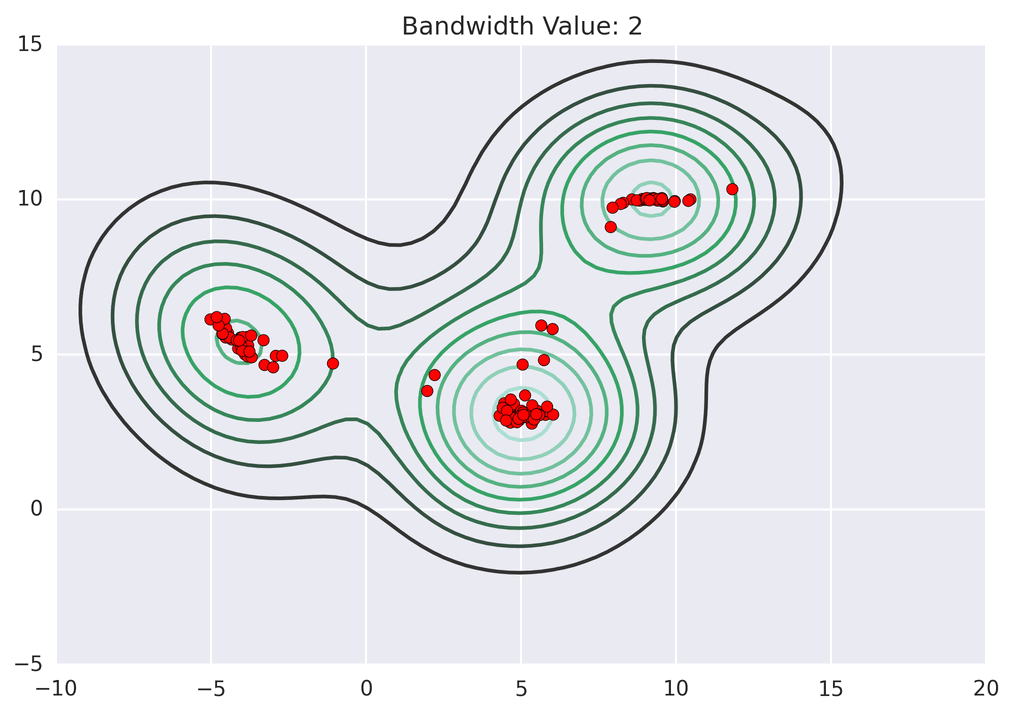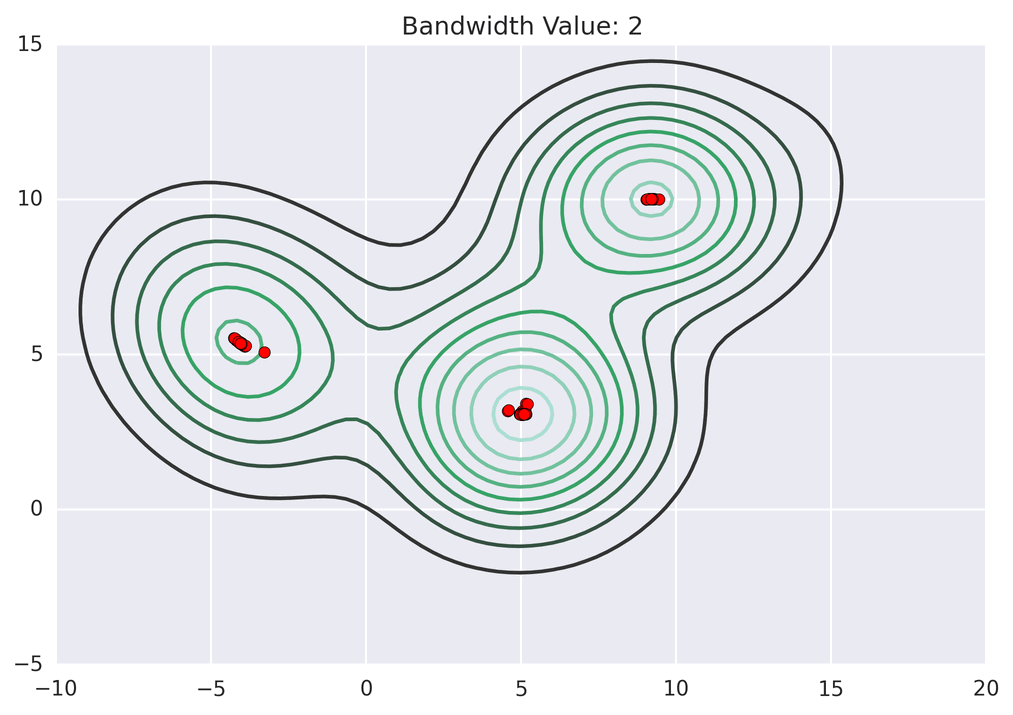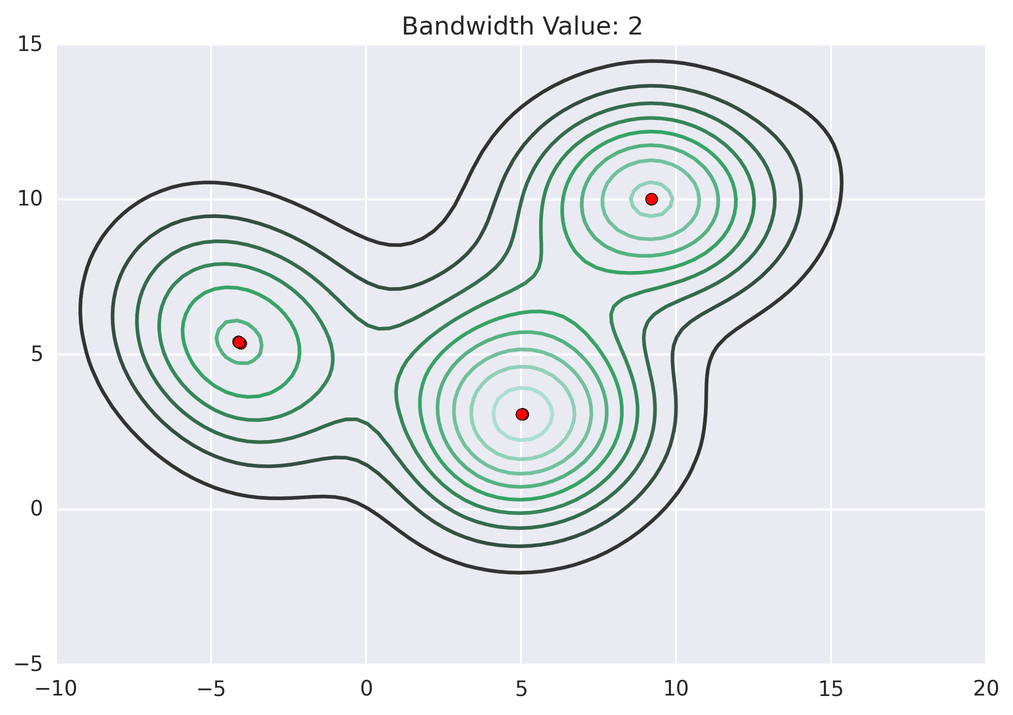
These GIFs from Matt Nedrich’s github illustrate the MeanShift process:

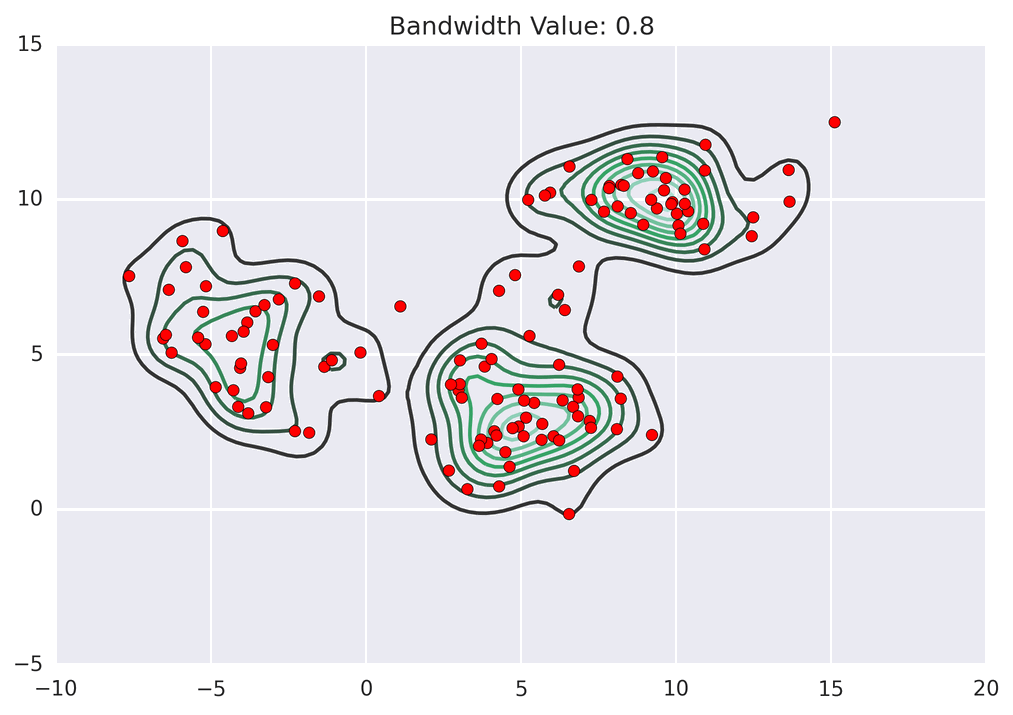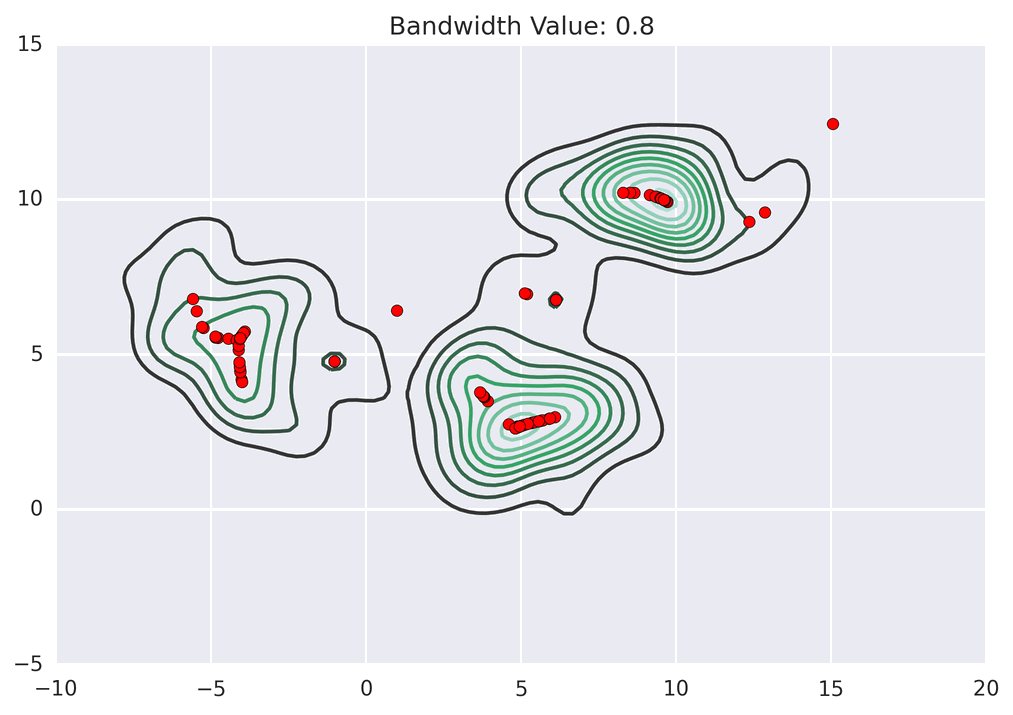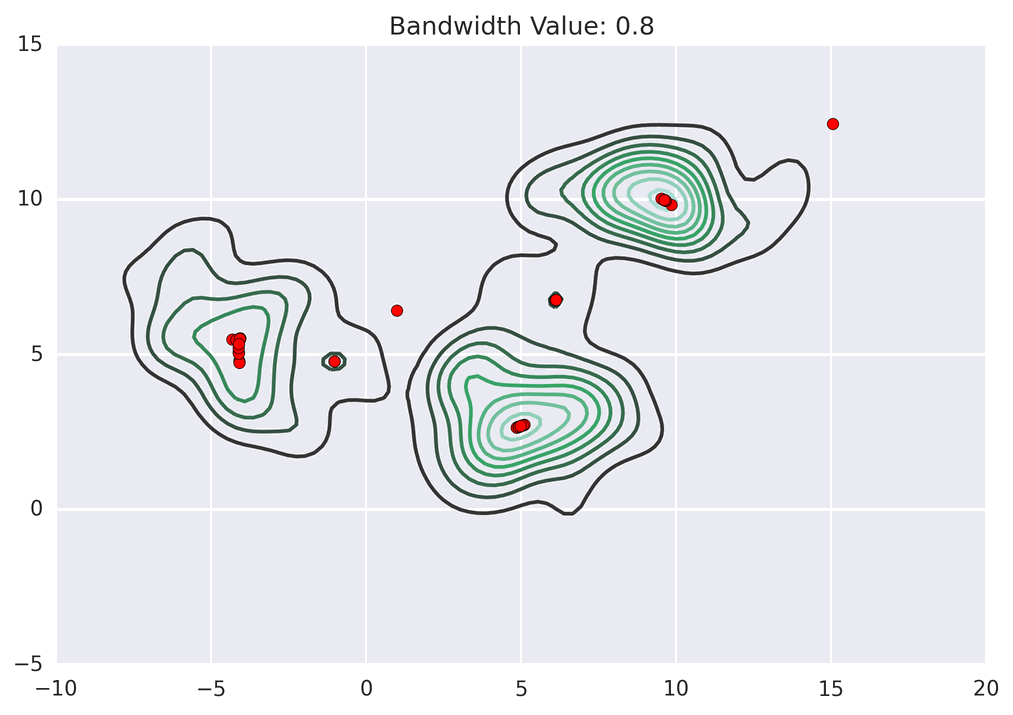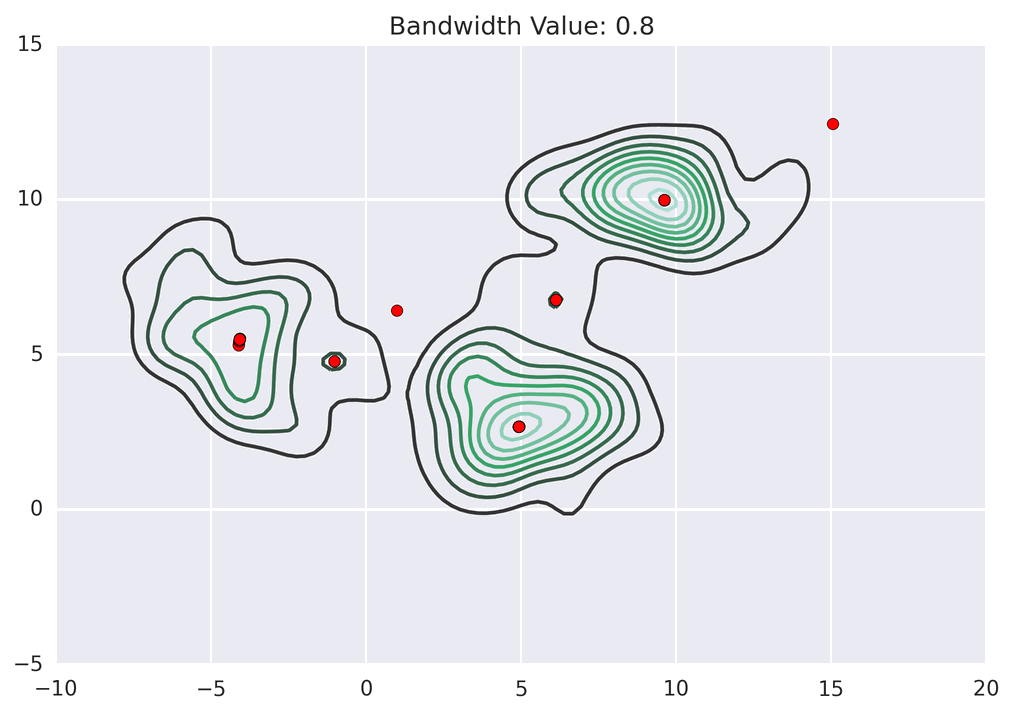
MeanShift clustering on a GPU
Since the algorithm outlined above is performed for each sample point i independently, they can all be computed in parallel. In fact, the MeanShift algorithm is quite computationally intensive – it is slow on a regular CPU. However, using a GPU it can be computed much more efficiently.
How much more efficiently? To test for yourself, download and install the MeanShiftGPU example program from the Sydney Informatics Hub GitHub (instructions are in the Readme). In this code, I have set the bandwidth parameter to 2.5x the smallest ‘expected’ standard deviation of the underlying distributions – but feel free to edit the code and try other values!
MeanShiftGPU uses the OpenCL framework to execute code in parallel. Though less well known, and supported, than NVIDIA’s CUDA, the open-source OpenCL has one distinct advantage: it can run on almost any processors, including CPUs. This is great because it means I can test GPU parallelism on my laptop (which has no NVIDIA cards), and also compare directly against the CPU’s performance on the same code.
MeanShiftGPU will first query your system to find available OpenCL devices, and ask you which to use. On my Macbook I have three options:
The following devices are available for use:
0) Intel Inc. : Intel(R) HD Graphics 630
1) AMD : AMD Radeon Pro 560 Compute Engine
2) Intel : Intel(R) Core(TM) i7-7920HQ CPU @ 3.10GHz
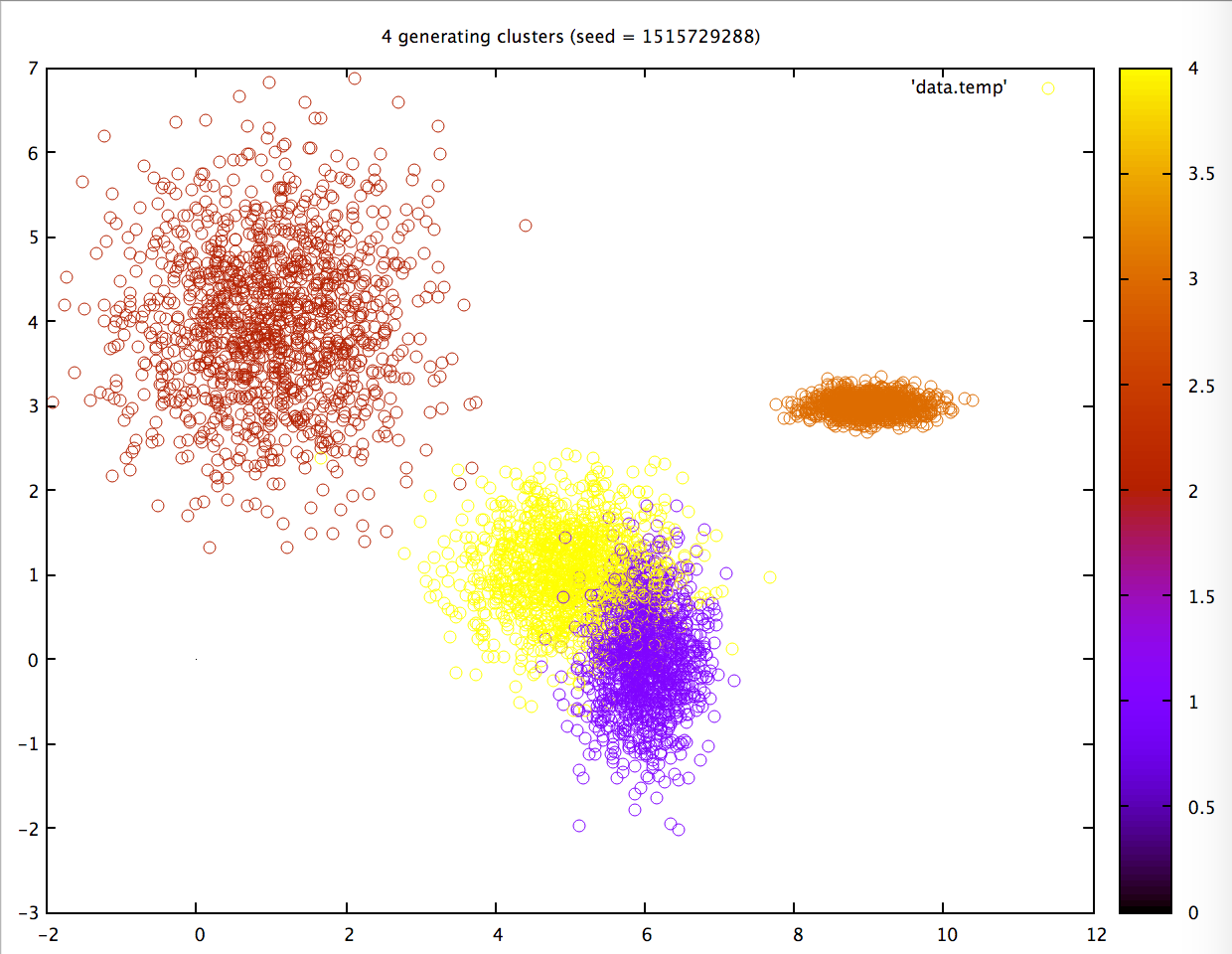
The first 2 are my Intel on-board graphics chip (actually on the CPU chip!), and my external AMD graphics card; the latter being the ‘serious’ GPU chip in my system. The 3rd option is my actual CPU. Running the program on my AMD chip, I get the following results:
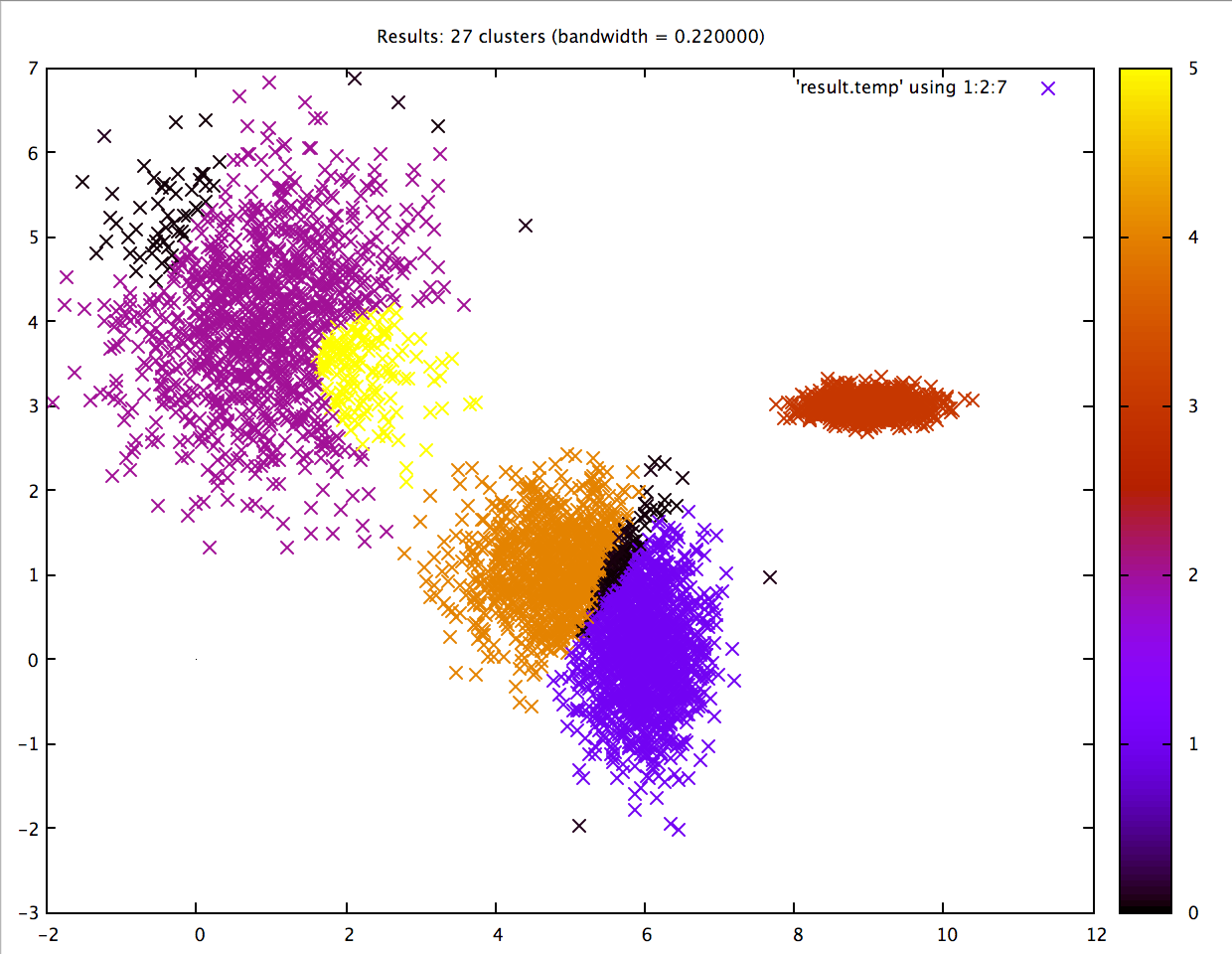
Not bad! Note how the two lower central clusters are nicely differentiated, but the upper left cluster is divided into several.3 The whole computation took less than 2 seconds:
100 Iterations on 20 work groups: Mean shifted 5120 points
Wall time 1.381684s
On the CPU, however, the same computation (seed = 1515729288) is much slower, 17x!
100 Iterations on 40 work groups: Mean shifted 5120 points
Wall time 24.290108s
Run MeanShiftGPU a couple of times on your systems, using whichever graphics processors the program finds. How does it perform? Then compare a run on your CPU. To see how the time taken scales with the size of the problem, I tested a few different sized datasets:
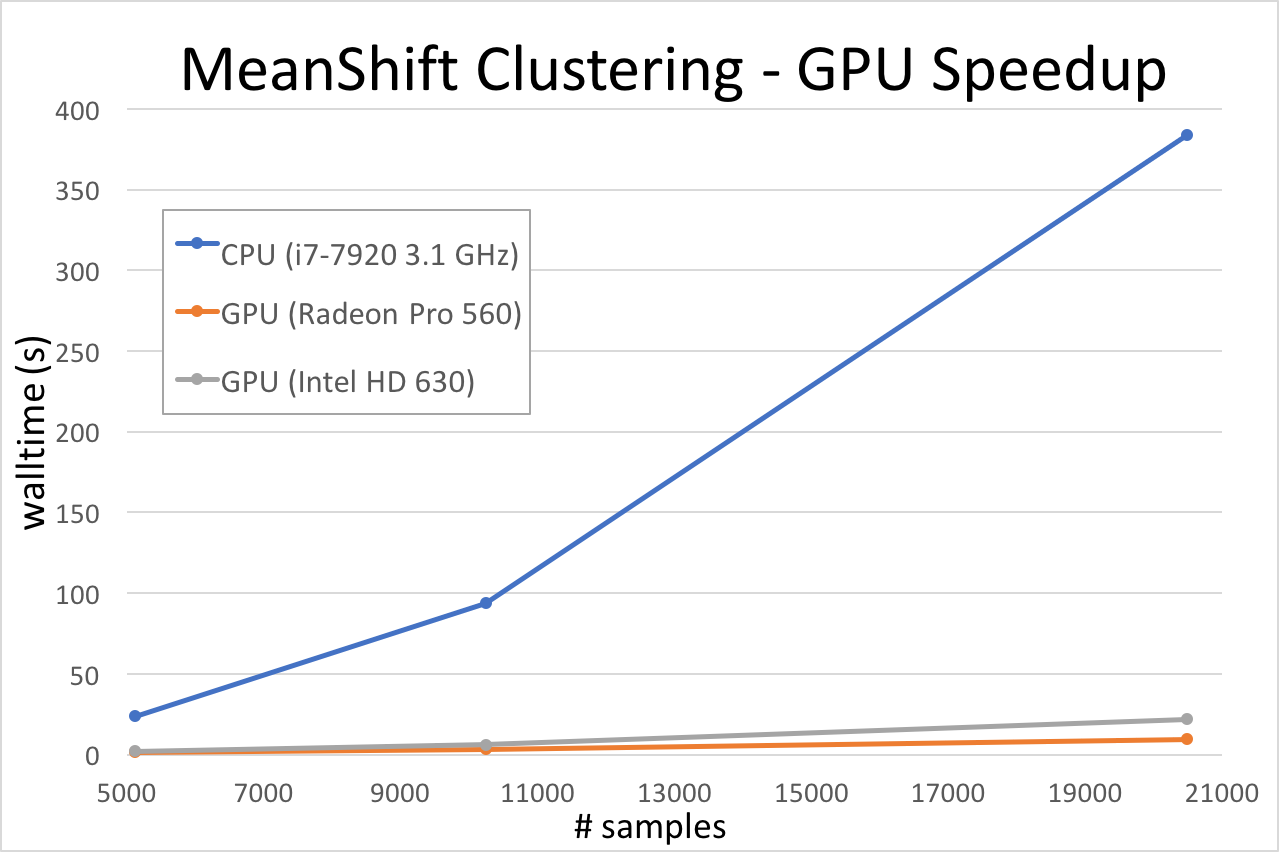
And now you can see why everyone is always talking about GPUs! When you have a massively – embarrassingly – parallelisable algorithm, definitely consider utilising a GPU. Artemis’s 2018 Phase-3 upgrade just added 28 NVIDIA Volta V100 GPGPUs with both CUDA and Tensor Cores — so it’s a great time to learn how to GPU. Keep a look out for SIH’s upcoming GPU course and, in the meantime, have a look at the online resources like:
- NVIDIA’s online training
- Lynda.com’s parallel Python/NumbaPro/PyCUDA/PyOpenCl course
- MATLAB’s GPU support in its Parallel Computing Toolbox
- This example has been adapted from two blog posts at Atomic Object: ‘Mean Shift Clustering’, and OpenCl for MacOSX. [return]
- In the sense that we needn’t assume anything about the shape of the data distributions. The algorithm itself requires at least one parameter, to define the neighbourhood function. [return]
- We can try different values of the bandwidth parameter to see if a better clustering can be found. The MeanShiftGPU currently sets the bandwidth automatically – to 2.5x the smallest ‘expected’ standard deviation of the underlying distributions – but it shouldn’t be hard to edit the source code to pick another value, or to make bandwidth a program argument. [return]